Section: New Results
Social-sparsity brain decoders: faster spatial sparsity
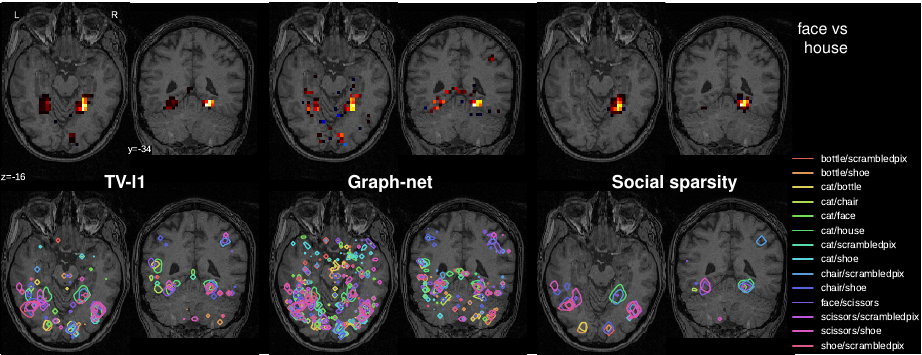
Spatially-sparse predictors are good models for brain decoding: they give accurate predictions and their weight maps are interpretable as they focus on a small number of regions. However, the state of the art, based on total variation or graph-net, is computationally costly. Here we introduce sparsity in the local neighborhood of each voxel with social-sparsity, a structured shrinkage operator. We find that, on brain imaging classification problems, social-sparsity performs almost as well as total-variation models and better than graph-net, for a fraction of the computational cost. It also very clearly outlines predictive regions. We give details of the model and the algorithm.
|
See Fig. 5 for an illustration and [32] for more information.